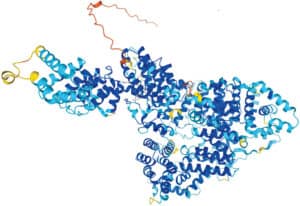
Researchers have said that the publication of this database, which is free to use, will help in combating malaria, antibiotic resistance, and plastic waste, as well as potentially speeding up the discovery of new drugs.
DeepMind has announced a huge expansion of its AlphaFold protein structure database, increasing the amount of 3D structures available from one million to over 200 million, which comprises almost all cataloged proteins known to science.
The project was started 18 months ago, although it was first conceptualized by DeepMind in 2016. It was considered to be one of the best ways for scientists to solve the 50 year old problem of protein folding, which is the identification of each protein’s unique structure.
SEE ALSO: Intel Launches Second-Generation Deep Learning Processors
“As someone who’s been in genomics and computational biology since the 1990s, I’ve seen many of these moments come where you can sense the landscape shifting under you and the provision of new resources, and this has been one of the fastest,” said joint director of EMBL’s European Bioinformatics Institute, Ewan Birney, to New Scientist. “I mean, two years ago, we just simply did not realize that this was feasible.”
Researchers have said that the publication of this database, which is free to use, will help in combating malaria, antibiotic resistance and plastic waste, as well as potentially speeding up the discovery of new drugs. DeepMind says it is proof that AI can have a positive impact on science.
DeepMind’s AlphaFold team worked with CASP (Critical Assessment of protein Structure Prediction), a community forum established in 1994 with the goal of accurately predicting protein structures. AlphaFold used their and other teams research to influence the model for the database, alongside participating in the annual protein structure prediction challenge in 2018, in which the AI came first.
What’s remarkable, for those involved, has been the speed at which AlphaFold has become superior to those with decades of experience in the field.
“AlphaFold is the singular and momentous advance in life science that demonstrates the power of AI,” said Eric Topol, Founder and Director of the Scripps Research Translational Institute. “Determining the 3D structure of a protein used to take many months or years, it now takes seconds. AlphaFold has already accelerated and enabled massive discoveries, including cracking the structure of the nuclear pore complex. And with this new addition of structures illuminating nearly the entire protein universe, we can expect more biological mysteries to be solved each day.”
This is not the first or the last time AI will be involved in the pursuit of cataloging and predicting scientific nodes and phenomenon. When the AI DeepMind and other organizations deal in, deep learning, was first being presented as a potential solution for scientific problems, it was as a tool to accelerate the massive computation needs for large projects such as protein folding.
“It’s the most complex thing we’ve ever done. This is the reason I started DeepMind,” said DeepMind co-founder and CEO, Demis Hassabis, to MIT Technology Review. “In fact, it’s why I’ve worked my whole career in AI.”